Homework 05: Segmentation for Connectomics
In this homework assignment, you'll have the opportunity to work on one of the most challenging problems in neuroscience: the automatic segmentation of neuronal processes.
For this assignment, record your responses to the following activities in the
README.md file in the homework05 folder of your assignments GitLab
repository and push your work by 11:59 PM Wednesday, November 13.
Activity 0: Branching
As discussed in class, each homework assignment must be completed in its own git branch; this will allow you to separate the work of each assignment and for you to use the merge request workflow.
To create a homework05 branch in your local repository, follow the
instructions below:
$ cd path/to/cse-40171-fa19-assignments # Go to assignments repository $ git remote add upstream https://gitlab.com/wscheirer/cse-40171-fa19-assignments # Switch back over to the main class repository $ git fetch upstream # Toggle the upstream branch $ git pull upstream master # Pull the files for homework05 $ git checkout -b homework05 # Create homework05 branch and check it out $ cd homework05 # Go into homework05 folder
Once these commands have been successfully performed, you are now ready to add, commit, and push any work required for this assignment.
Activity 1: Segment an APEX-labeled EM Volume of Retinal Neurons (100 Points)
In class we have been learning about the imaging techniques that neuroscientists use to study the connectivity between neurons in the nervous system. Access to a connectome has potentially important ramifications for AI, as it provides a roadmap to neural circuits that can guide the wiring of artificial neural networks. Electron Microscopy is one of the most powerful imaging modalities, so much so that it can resolve the synaptic connections between cells. Because imaged volumes of brain tissue can be terabytes or petabytes in size, with each individual image an exceptionally information-rich piece of data, it is not possible to parse all of it by hand. Thus we turn to the use of computer vision for automatic segmentation. The general problem is known as "dense" segmentation, in that all membranes (the thin boundaries between cells) should be separated from the background. Understanding the scale at which this takes place is important. The resolution of EM can be so high that we do not see cell bodies, just the intricate wiring of the axons and dendrites from the cells. Depending on what we want to study, we can make the computer vision problem a little bit easier with some help from our biologist friends.
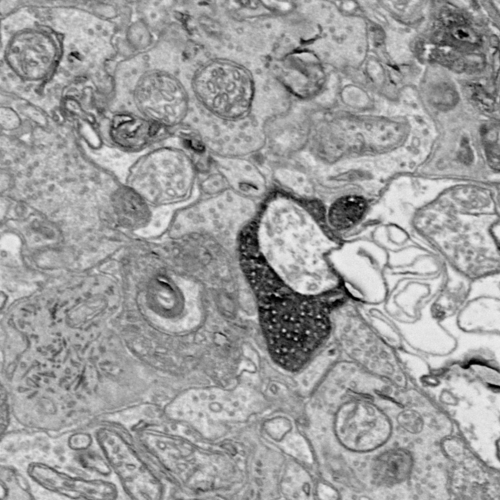
The tissue in the image above has been prepared with a special peroxidase-based marker called APEX. It is a genetically targeted marker that can increase the contrast of certain cell types. The use of APEX allows neuroscientists to prepare volumes that make the job of the computer scientist easier — an automatic segmentation approach just needs to isolate the darkest pixel clusters in each image. However, this changes the problem from dense segmentation to sparse segmentation, where only neural circuits composed of known cell types can be reconstructed. Nonetheless, this is a good starting place for learning about connectomics segmentation.
Your task in this assignment is to implement a segmentation approach that can separate the APEX labeled processes from the background pixels. This means that the output of the tool should be segmentation masks that look similar to the corresponding ground-truth. You are free to use any approach that you'd like, including approaches we haven't discussed in class. The approach must be implemented in python (i.e., stay away from standalone applications for the processing of medical images like fiji and ilastik). Both learning-free and machine learning-based approaches are valid for this assignment.
The image data can be downloaded here. This stack of images is a sub-volume of the retina volume imaged by serial-scanning electron microscopy that is described in this paper. After unzipping the archive, you will find two directories. The original EM images are in the images/ directory. Ground-truth masks are in the ground-truth/ directory. These masks show the pixels that belong to all APEX-labeled processes in an image. Note that partial ground-truth has been provided — only 45 ground-truth images are available. Thus, you can treat images 000-0044 as training data (useful even if you are going for a learning-free approach, because a threshold may need to be tuned), and the rest as testing data. Also note that not all of the images have APEX-labeled processes. The ground-truth image is completely black for those instances.
Segment all of the provided images and push the images of your segmentation masks to your repository. Also push any code that you have developed. Your README.md should document the approach you used (citing any relevant sources), as well as any dependencies needed for installation. You will be assessed on the visual quality of the segmentations you produce on both the training and testing sets.
Feedback
If you have any questions, comments, or concerns regarding the course, please
provide your feedback at the end of your README.md.
Submission
To submit your assignment, please commit your work to the homework05 folder
of your homework05 branch in your assignment's GitLab repository:
$ cd path/to/cse-40171-fa19-assignments # Go to assignments repository $ git checkout master # Make sure we are in master branch $ git pull --rebase # Make sure we are up-to-date with GitLab $ git checkout -b homework05 # Create homework05 branch and check it out $ cd homework05 # Go to homework05 directory ... $ $EDITOR README.md # Edit appropriate README.md $ git add README.md # Mark changes for commit $ git commit -m "homework05: complete" # Record changes ... $ git push -u origin homework05 # Push branch to GitLab
Procedure for submitting your work: create a merge request by the process that is described here, but make sure to change the target branch from wscheirer/cse-40567-sp19-assignments to your personal fork's master branch so that your code is not visible to other students. Additionally, assign this merge request to our TA (sabraha2) and add wscheirer as an approver (so all class staff can track your submission).